import pandas as pd
import dabest
from palmerpenguins import load_penguinsPre-compiling numba functions for DABEST...Compiling numba functions: 100%|████████████████| 11/11 [00:00<00:00, 67.69it/s]Numba compilation complete!March 28, 2025
DABEST is a package that performs estimation statistics available on Python and R. With Jupyter Notebook you can try DABEST-Python.
Pre-compiling numba functions for DABEST...Compiling numba functions: 100%|████████████████| 11/11 [00:00<00:00, 67.69it/s]Numba compilation complete!| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | male | 2007 |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | female | 2007 |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | female | 2007 |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN | 2007 |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | female | 2007 |
DABEST v2025.03.27
==================
Good evening!
The current time is Thu Mar 27 23:02:20 2025.
The unpaired mean difference between Adelie and Chinstrap is 10.0 [95%CI 9.14, 11.0].
The p-value of the two-sided permutation t-test is 0.0, calculated for legacy purposes only.
The unpaired mean difference between Adelie and Gentoo is 8.71 [95%CI 8.02, 9.42].
The p-value of the two-sided permutation t-test is 0.0, calculated for legacy purposes only.
5000 bootstrap samples were taken; the confidence interval is bias-corrected and accelerated.
Any p-value reported is the probability of observing theeffect size (or greater),
assuming the null hypothesis of zero difference is true.
For each p-value, 5000 reshuffles of the control and test labels were performed.
To get the results of all valid statistical tests, use `.mean_diff.statistical_tests`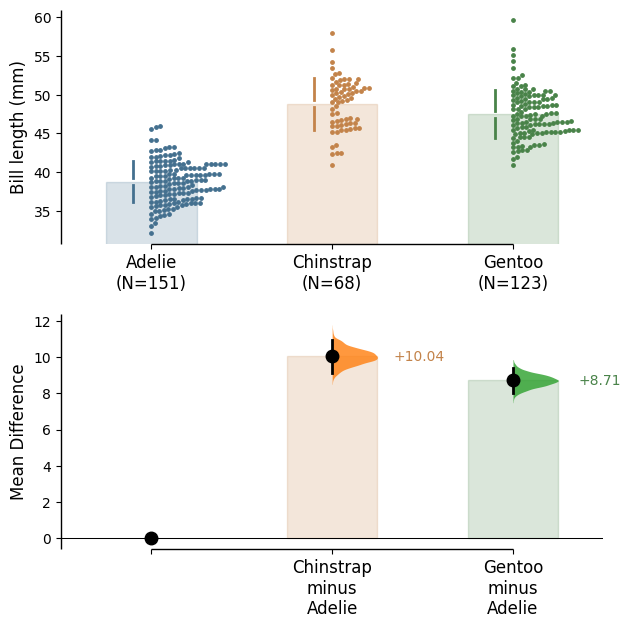
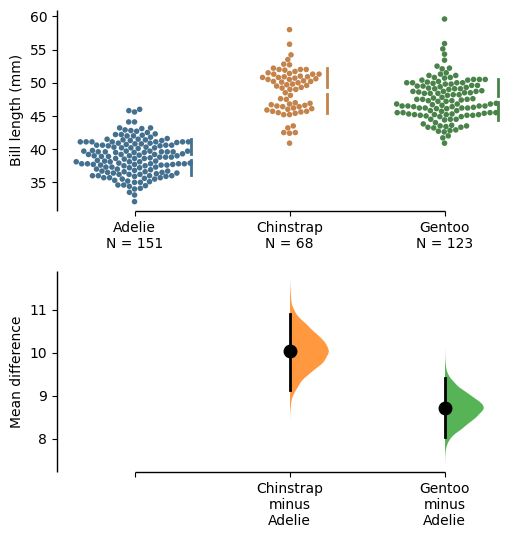
The DABEST library has also been developed into a web application at estimationstats.com