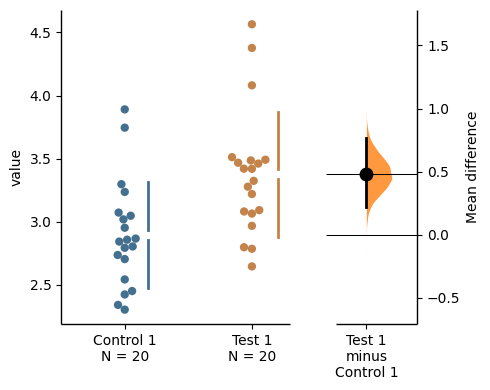
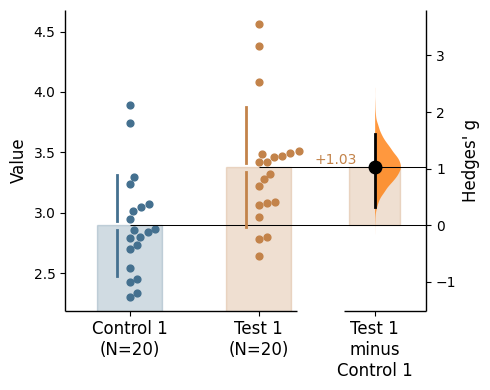
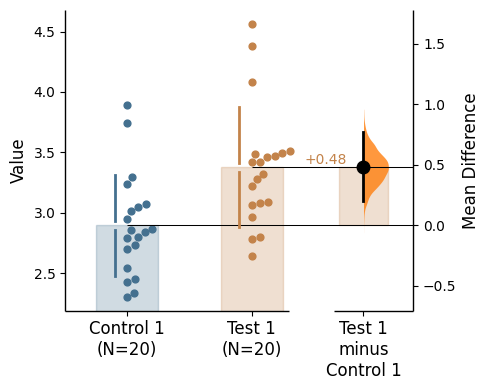
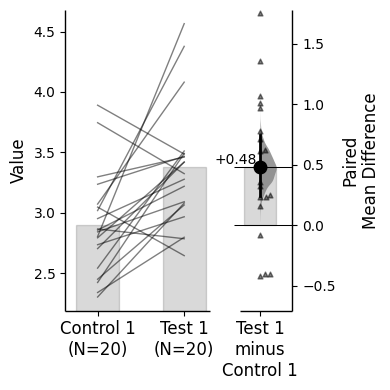
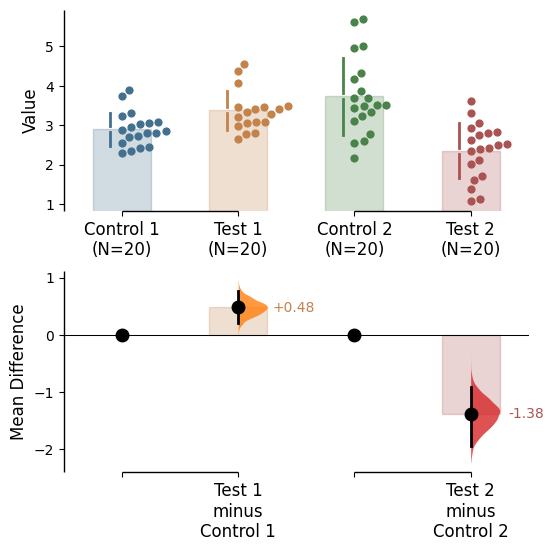
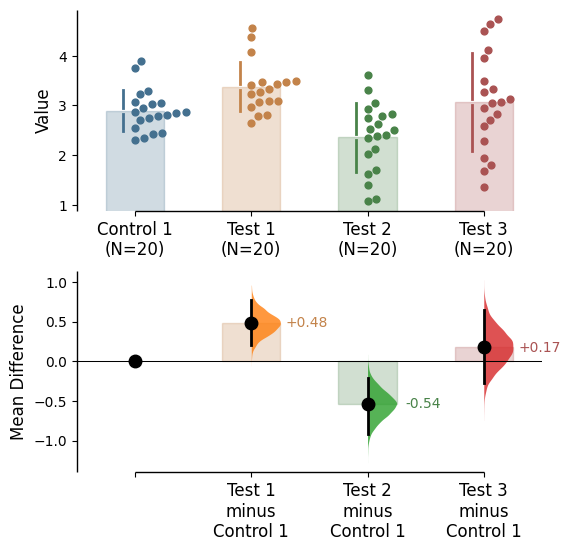
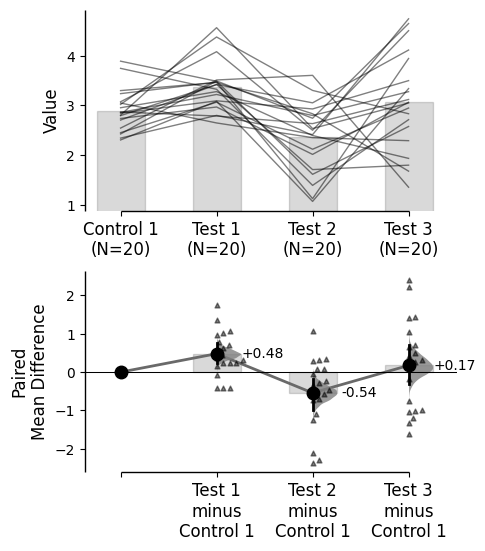
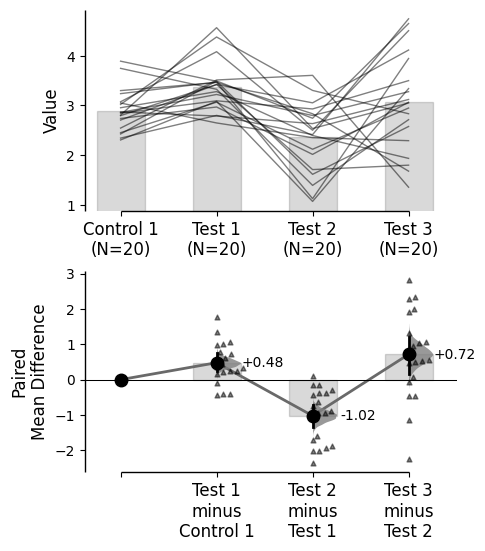
{'alpha': 0.05,
'bca_high': 0.2413346581369784,
'bca_interval_idx': (109, 4858),
'bca_low': -0.7818088458343655,
'bec_bca_high': 0.5352403905584314,
'bec_bca_interval_idx': (130, 4880),
'bec_bca_low': -0.4982839949134528,
'bec_bootstraps': array([-0.48953946, -0.18565285, -0.23896785, ..., -0.55130928,
0.16037238, -0.07364879]),
'bec_difference': 0.0,
'bec_pct_high': 0.5280564736117328,
'bec_pct_interval_idx': (125, 4875),
'bec_pct_low': -0.5041777340626885,
'bootstraps': array([-0.23923425, -0.66013733, -0.42672232, ..., -0.33191074,
-0.16543251, -0.34179536]),
'ci': 95,
'difference': -0.25315417702752846,
'effect_size': 'mean difference',
'is_paired': None,
'is_proportional': False,
'pct_high': 0.25135646125431527,
'pct_interval_idx': (125, 4875),
'pct_low': -0.763588353717278,
'permutation_count': 5000,
'permutations': array([ 0.17221029, 0.03112419, -0.13911387, ..., -0.38007941,
0.30261507, -0.09073054]),
'permutations_var': array([0.07201642, 0.07251104, 0.07219407, ..., 0.07003705, 0.07094885,
0.07238581]),
'proportional_difference': nan,
'pvalue_brunner_munzel': nan,
'pvalue_kruskal': nan,
'pvalue_mann_whitney': 0.5201446121616038,
'pvalue_mcnemar': nan,
'pvalue_paired_students_t': nan,
'pvalue_permutation': 0.3484,
'pvalue_students_t': 0.34743913903372836,
'pvalue_welch': 0.3474493875548964,
'pvalue_wilcoxon': nan,
'random_seed': 12345,
'resamples': 5000,
'statistic_brunner_munzel': nan,
'statistic_kruskal': nan,
'statistic_mann_whitney': 494.0,
'statistic_mcnemar': nan,
'statistic_paired_students_t': nan,
'statistic_students_t': 0.9472545159069105,
'statistic_welch': 0.9472545159069105,
'statistic_wilcoxon': nan}