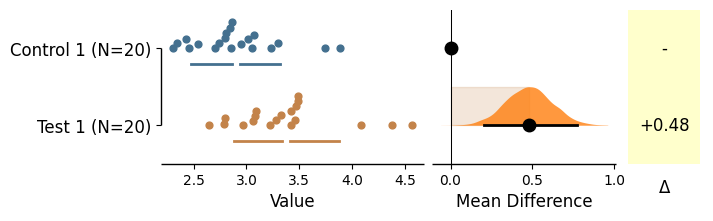
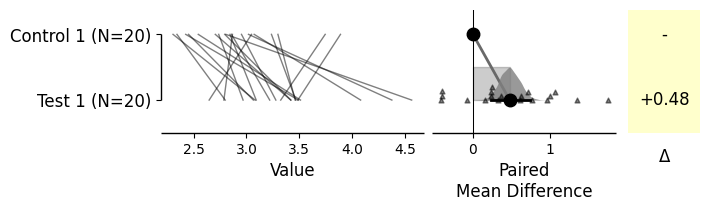
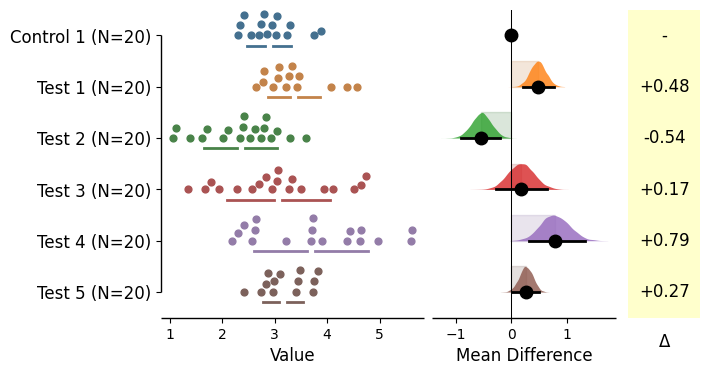
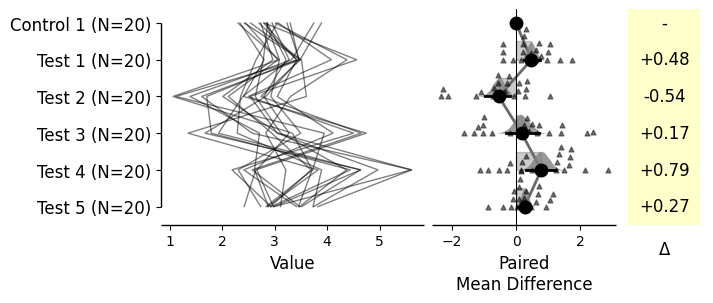
In DABEST v2025.03.27 , we introduce a new plotting orientation: horizontal plots .
To access this, provide horizontal=True to the .plot() method.
Load libraries
import numpy as npimport pandas as pdimport dabestprint ("We're using DABEST v {} " .format (dabest.__version__))
Pre-compiling numba functions for DABEST...
Compiling numba functions: 100%|██████████| 11/11 [00:00<00:00, 30.36it/s]
Numba compilation complete!
We're using DABEST v2025.03.27
Creating a demo dataset
from scipy.stats import norm # Used in generation of populations. 9999 ) # Fix the seed to ensure reproducibility of results. = 20 # The number of samples taken from each population # Create samples = norm.rvs(loc= 3 , scale= 0.4 , size= Ns)= norm.rvs(loc= 3.5 , scale= 0.75 , size= Ns)= norm.rvs(loc= 3.25 , scale= 0.4 , size= Ns)= norm.rvs(loc= 3.5 , scale= 0.5 , size= Ns)= norm.rvs(loc= 2.5 , scale= 0.6 , size= Ns)= norm.rvs(loc= 3 , scale= 0.75 , size= Ns)= norm.rvs(loc= 3.5 , scale= 0.75 , size= Ns)= norm.rvs(loc= 3.25 , scale= 0.4 , size= Ns)= norm.rvs(loc= 3.25 , scale= 0.4 , size= Ns)# Add a `gender` column for coloring the data. = np.repeat('Female' , Ns/ 2 ).tolist()= np.repeat('Male' , Ns/ 2 ).tolist()= females + males# Add an `id` column for paired data plotting. = pd.Series(range (1 , Ns+ 1 ))# Combine samples and gender into a DataFrame. = pd.DataFrame({'Control 1' : c1, 'Test 1' : t1,'Control 2' : c2, 'Test 2' : t2,'Control 3' : c3, 'Test 3' : t3,'Test 4' : t4, 'Test 5' : t5, 'Test 6' : t6,'Gender' : gender, 'ID' : id_col
Generating two-group plots
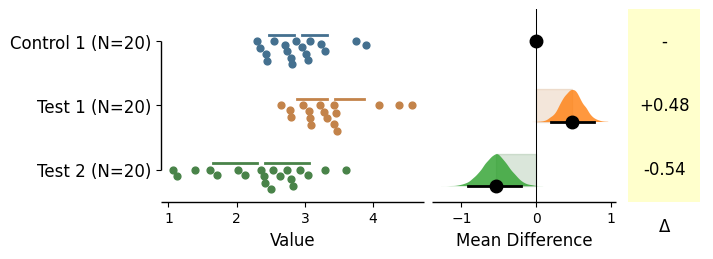
= dabest.load(df, idx= ("Control 1" , "Test 1" ))= True ); = dabest.load(df, idx= ("Control 1" , "Test 1" ), paired= 'baseline' , id_col= 'ID' )= True );
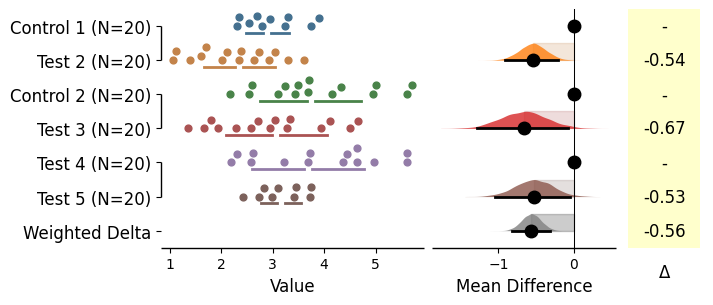
Generating shared-control and repeated-measures plots
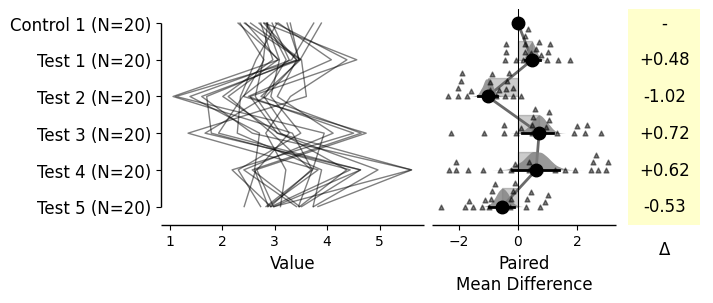
= dabest.load(df, idx= ("Control 1" , "Test 1" , "Test 2" , "Test 3" , "Test 4" , "Test 5" ))= True ); = dabest.load(df, idx= ("Control 1" , "Test 1" , "Test 2" , "Test 3" , "Test 4" , "Test 5" ), paired= 'baseline' , id_col= 'ID' ) = True ); = dabest.load(df, idx= ("Control 1" , "Test 1" , "Test 2" , "Test 3" , "Test 4" , "Test 5" ), paired= 'sequential' , id_col= 'ID' ) = True );
Generating multi-group plots
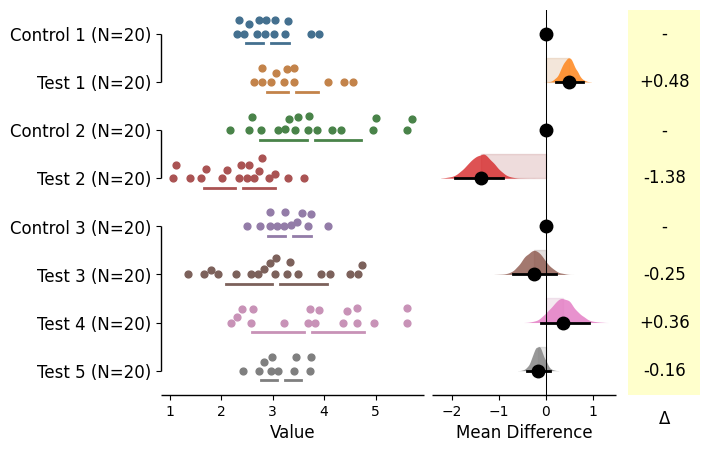
= dabest.load(df, idx= (("Control 1" , "Test 1" ),("Control 2" , "Test 2" ),("Control 3" , "Test 3" , "Test 4" , "Test 5" )))= True );
Generating proportion plots
def create_demo_prop_dataset(seed= 9999 , N= 40 ):import numpy as npimport pandas as pd9999 ) # Fix the seed to ensure reproducibility of results. # Create samples = 1 = np.random.binomial(n, 0.2 , size= N)= np.random.binomial(n, 0.2 , size= N)= np.random.binomial(n, 0.8 , size= N)= np.random.binomial(n, 0.6 , size= N)= np.random.binomial(n, 0.2 , size= N)= np.random.binomial(n, 0.3 , size= N)= np.random.binomial(n, 0.4 , size= N)= np.random.binomial(n, 0.5 , size= N)= np.random.binomial(n, 0.6 , size= N)= np.ones(N)= np.zeros(N)= np.zeros(N)# Add a `gender` column for coloring the data. = np.repeat('Female' , N / 2 ).tolist()= np.repeat('Male' , N / 2 ).tolist()= females + males# Add an `id` column for paired data plotting. = pd.Series(range (1 , N + 1 ))# Combine samples and gender into a DataFrame. = pd.DataFrame({'Control 1' : c1, 'Test 1' : t1,'Control 2' : c2, 'Test 2' : t2,'Control 3' : c3, 'Test 3' : t3,'Test 4' : t4, 'Test 5' : t5, 'Test 6' : t6,'Test 7' : t7, 'Test 8' : t8, 'Test 9' : t9,'Gender' : gender, 'ID' : id_colreturn df= create_demo_prop_dataset()
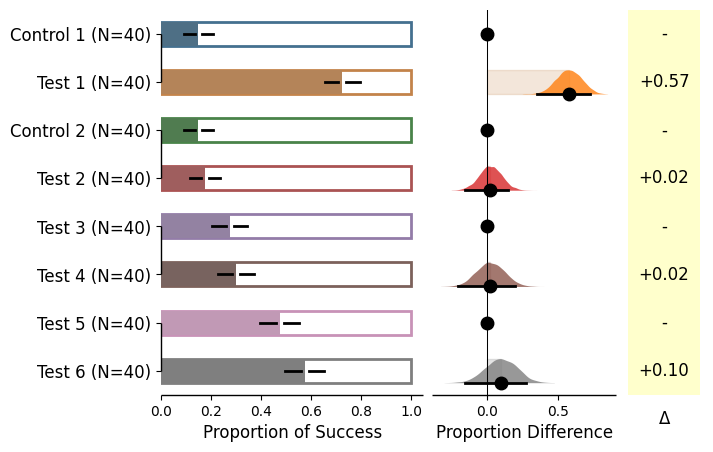
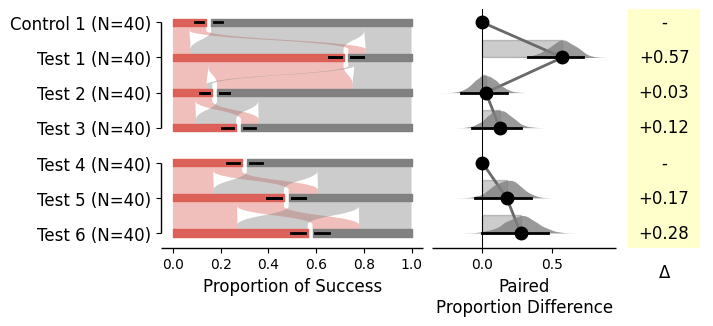
= dabest.load(df_prop, idx= (("Control 1" , "Test 1" ), ("Control 2" , "Test 2" ), ("Test 3" , "Test 4" ), ("Test 5" , "Test 6" )), proportional= True )= True ); = dabest.load(df_prop, idx= ((("Control 1" , "Test 1" ,"Test 2" , "Test 3" ),("Test 4" , "Test 5" , "Test 6" ))),proportional= True , paired= "baseline" , id_col= "ID" )= True );
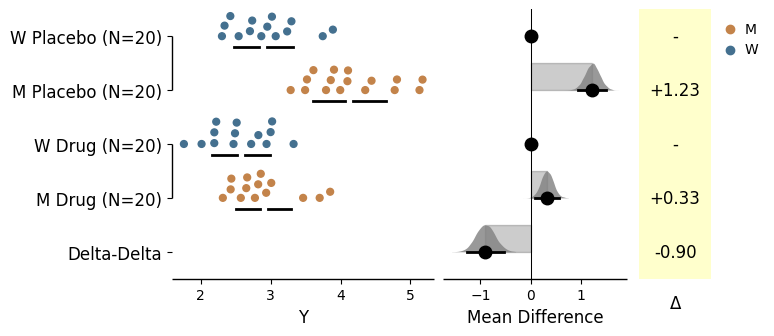
Generating delta-delta plots
from scipy.stats import norm # Used in generation of populations. 9999 ) # Fix the seed to ensure reproducibility of results. # Create samples = 20 = norm.rvs(loc= 3 , scale= 0.4 , size= N* 4 )2 * N] = y[N:2 * N]+ 1 2 * N:3 * N] = y[2 * N:3 * N]- 0.5 # Add a `Treatment` column = np.repeat('Placebo' , N* 2 ).tolist()= np.repeat('Drug' , N* 2 ).tolist()= t1 + t2 # Add a `Rep` column as the first variable for the 2 replicates of experiments done = []for i in range (N* 2 ):'Rep1' )'Rep2' )# Add a `Genotype` column as the second variable = np.repeat('W' , N).tolist()= np.repeat('M' , N).tolist()= np.repeat('W' , N).tolist()= np.repeat('M' , N).tolist()= wt + mt + wt2 + mt2# Add an `id` column for paired data plotting. id = list (range (0 , N* 2 ))= id + id # Combine all columns into a DataFrame. = pd.DataFrame({'ID' : id_col,'Rep' : rep,'Genotype' : genotype, 'Treatment' : treatment,'Y' : y
= dabest.load(data = df_delta2, x = ["Genotype" , "Genotype" ], y = "Y" , delta2 = True , experiment = "Treatment" )= True );
Controlling aesthetics
As with the vertical plots, horizontal plots can be customized using the same options. Shown below are a few cases where the aesthetics are modified, added functionality, or just less intuitive.
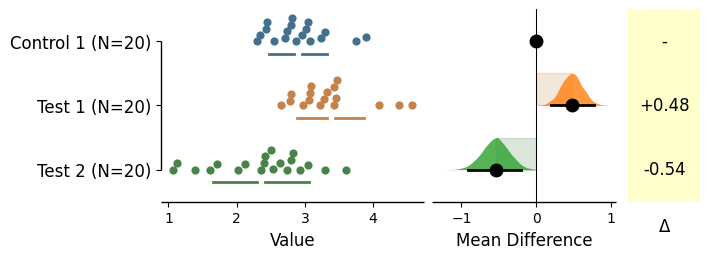
Swarm side
As with the vertical plots, you can specify the side of the swarms via swarm_side in the .plot() method.
In this case, swarm_side='left' would plot the swarms upwards, and swarm_side='right' would plot the swarms downwards.
Default is swarm_side='left'
= dabest.load(df, idx= ("Control 1" , "Test 1" , 'Test 2' ), resamples= 5000 )= 'left' , horizontal= True );
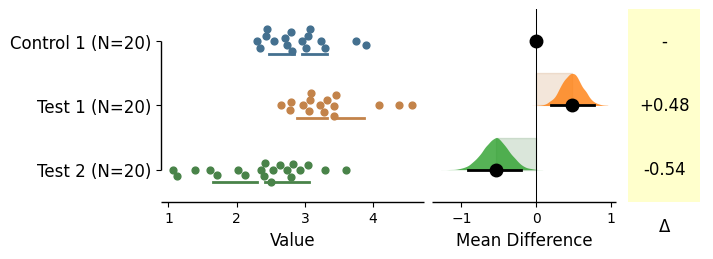
swarm_side='center'
= 'center' , horizontal= True );
swarm_side='right'
= 'right' , horizontal= True );
Table kwargs
The table axis can be customized using the horizontal_table_kwargs argument. A dict of keywords can be passed to customize the table.
If None, the following keywords are passed:
'show' - Whether to show the table. Default is True.'color' - The color of the table. Default is ‘yellow’.'alpha' - The transparency of the table. Default is 0.2.'fontsize' - The fontsize of the table. Default is 12.'text_color' - The color of the text in the table. Default is ‘black’.'text_units' - The units of the text in the table. Default is None.'control_marker' - The marker for the control group. Default is ‘-’.'fontsize_label' - The fontsize of the table x-label. Default is 12.'label' - The table x-label.
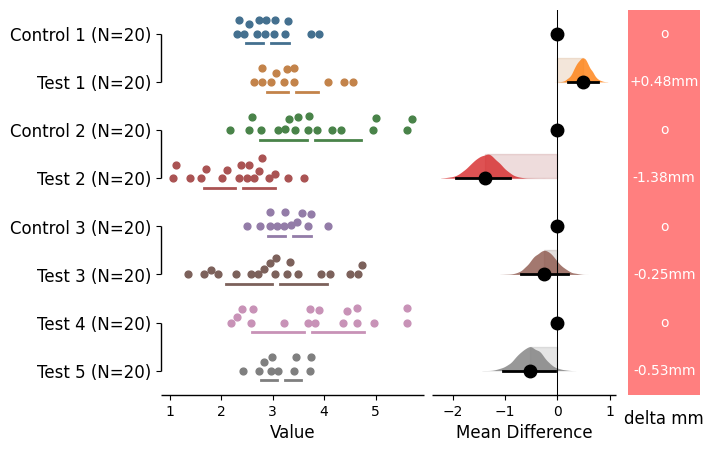
= dabest.load(df, idx= (("Control 1" , "Test 1" ),("Control 2" , "Test 2" ),("Control 3" , "Test 3" ),("Test 4" , "Test 5" )))= True , = {'color' : 'red' , 'alpha' : 0.5 , 'text_color' : 'white' ,'text_units' :'mm' , 'label' : 'delta mm' ,'control_marker' : 'o' ,;
The table axis can be hidden using the 'show':False in the horizontal_table_kwargs dict.
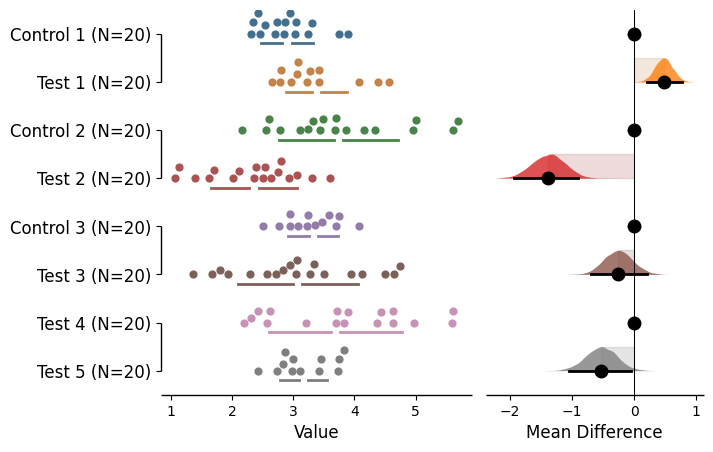
= dabest.load(df, idx= (("Control 1" , "Test 1" ),("Control 2" , "Test 2" ),("Control 3" , "Test 3" ),("Test 4" , "Test 5" )))= True , horizontal_table_kwargs= {'show' : False });
Gridkey
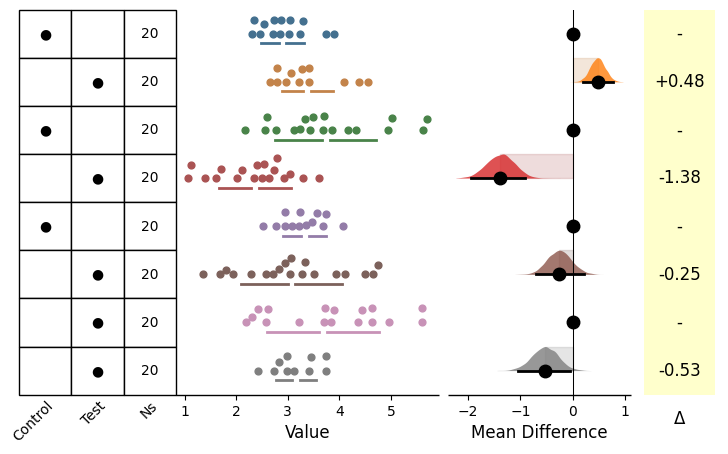
As with the vertical plots, you can utilise a gridkey table for representing the groupings. This can be reached via gridkey in the .plot() method.
You can either use gridkey='auto' to automatically generate the gridkey, or pass a list of indexes to represent the groupings (e.g., gridkey=['Control', 'Test']).
See the examples in the Plot Aesthetics Tutorial for more information with regards to kwargs.
= dabest.load(df, idx= (("Control 1" , "Test 1" ),("Control 2" , "Test 2" ),("Control 3" , "Test 3" ),("Test 4" , "Test 5" )))= True , gridkey= ['Control' , 'Test' ]);