import numpy as np
import pandas as pd
import dabest
print("We're using DABEST v{}".format(dabest.__version__))We're using DABEST v2024.03.29Since version 2023.02.14, DABEST also supports the calculation of delta-delta, an experimental function that facilitates the comparison between two bootstrapped effect sizes computed from two independent categorical variables.
Many experimental designs investigate the effects of two interacting independent variables on a dependent variable. The delta-delta effect size enables us distill the net effect of the two variables. To illustrate this, let’s explore the following problem.
Consider an experiment where we test the efficacy of a drug named Drug on a disease-causing mutation M based on disease metric Y. The greater the value Y has, the more severe the disease phenotype is. Phenotype Y has been shown to be caused by a gain-of-function mutation M, so we expect a difference between wild type (W) subjects and mutant subjects (M). Now, we want to know whether this effect is ameliorated by the administration of Drug treatment. We also administer a placebo as a control. In theory, we only expect Drug to have an effect on the M group, although in practice, many drugs have non-specific effects on healthy populations too.
Effectively, we have four groups of subjects for comparison.
| Wildtype | Mutant | |
|---|---|---|
| Drug | XD, W | XD, M |
| Placebo | XP, W | XP, M |
There are two Treatment conditions, Placebo (control group) and Drug (test group). There are two Genotype: W (wild type population) and M (mutant population). Additionally, each experiment was conducted twice (Rep1 and Rep2). We will perform several analyses to visualise these differences in a simulated dataset.
from scipy.stats import norm # Used in generation of populations.
np.random.seed(9999) # Fix the seed to ensure reproducibility of results.
# Create samples
N = 20
y = norm.rvs(loc=3, scale=0.4, size=N*4)
y[N:2*N] = y[N:2*N]+1
y[2*N:3*N] = y[2*N:3*N]-0.5
# Add a `Treatment` column
t1 = np.repeat('Placebo', N*2).tolist()
t2 = np.repeat('Drug', N*2).tolist()
treatment = t1 + t2
# Add a `Rep` column as the first variable for the 2 replicates of experiments done
rep = []
for i in range(N*2):
rep.append('Rep1')
rep.append('Rep2')
# Add a `Genotype` column as the second variable
wt = np.repeat('W', N).tolist()
mt = np.repeat('M', N).tolist()
wt2 = np.repeat('W', N).tolist()
mt2 = np.repeat('M', N).tolist()
genotype = wt + mt + wt2 + mt2
# Add an `id` column for paired data plotting.
id = list(range(0, N*2))
id_col = id + id
# Combine all columns into a DataFrame.
df_delta2 = pd.DataFrame({'ID' : id_col,
'Rep' : rep,
'Genotype' : genotype,
'Treatment': treatment,
'Y' : y
})To create a delta-delta plot, you simply need to set delta2=True in the dabest.load() function. However, in this case,x needs to be declared as a list consisting of 2 elements, unlike most cases where it is a single element. The first element in x will represent the variable plotted along the horizontal axis, and the second one will determine the color of dots for scattered plots or the color of lines for slope graphs. We use the experiment input to specify the grouping of the data.
The above function creates the following object:
DABEST v2024.03.29
==================
Good afternoon!
The current time is Tue Mar 19 15:44:01 2024.
Effect size(s) with 95% confidence intervals will be computed for:
1. M Placebo minus W Placebo
2. M Drug minus W Drug
3. Drug minus Placebo (only for mean difference)
5000 resamples will be used to generate the effect size bootstraps.We can quickly check out the effect sizes:
DABEST v2024.03.29
==================
Good afternoon!
The current time is Tue Mar 19 15:44:04 2024.
The unpaired mean difference between W Placebo and M Placebo is 1.23 [95%CI 0.948, 1.52].
The p-value of the two-sided permutation t-test is 0.0, calculated for legacy purposes only.
The unpaired mean difference between W Drug and M Drug is 0.326 [95%CI 0.0934, 0.584].
The p-value of the two-sided permutation t-test is 0.0122, calculated for legacy purposes only.
The delta-delta between Placebo and Drug is -0.903 [95%CI -1.27, -0.522].
The p-value of the two-sided permutation t-test is 0.0, calculated for legacy purposes only.
5000 bootstrap samples were taken; the confidence interval is bias-corrected and accelerated.
Any p-value reported is the probability of observing the effect size (or greater),
assuming the null hypothesis of zero difference is true.
For each p-value, 5000 reshuffles of the control and test labels were performed.
To get the results of all valid statistical tests, use `.mean_diff.statistical_tests`In the above plot, the horizontal axis represents the Genotype condition and the dot colour is also specified by Genotype. The left pair of scattered plots is based on the Placebo group while the right pair is based on the Drug group. The bottom left axis contains the two primary deltas: the Placebo delta and the Drug delta. We can easily see that when only the placebo was administered, the mutant phenotype is around 1.23 [95%CI 0.948, 1.52]. This difference was shrunken to around 0.326 [95%CI 0.0934, 0.584] when the drug was administered. This gives us some indication that the drug is effective in amiliorating the disease phenotype. Since the Drug did not completely eliminate the mutant phenotype, we have to calculate how much net effect the drug had. This is where delta-delta comes in. We use the Placebo delta as a reference for how much the mutant phenotype is supposed to be, and we subtract the Drug delta from it. The bootstrapped mean differences (delta-delta) between the Placebo and Drug group are plotted at the right bottom with a separate y-axis from other bootstrap plots. This effect size, at about -0.903 [95%CI -1.28, -0.513], is the net effect size of the drug treatment. That is to say that treatment with drug A reduced disease phenotype by 0.903.
The mean difference between mutants and wild types given the placebo treatment is:
\(\Delta_{1} = \overline{X}_{P, M} - \overline{X}_{P, W}\)
The mean difference between mutants and wild types given the drug treatment is:
\(\Delta_{2} = \overline{X}_{D, M} - \overline{X}_{D, W}\)
The net effect of the drug on mutants is:
\(\Delta_{\Delta} = \Delta_{2} - \Delta_{1}\)
where \(\overline{X}\) is the sample mean, \(\Delta\) is the mean difference.
In the example above, we used the convention of test - control but you can manipulate the orders of the experiment groups as well as the horizontal axis variable by setting the paremeters experiment_label and x1_level.
The delta-delta function also supports paired data, providing a useful alternative visualization of the data. Assuming that the placebo and drug treatment were administered to the same subjects, our data is paired between the treatment conditions. We can specify this by using Treatment as x and Genotype as experiment, and we further specify that id_col is ID, linking data from the same subject with each other. Since we have conducted two replicates of the experiments, we can also colour the slope lines according to Rep.
paired_delta2 = dabest.load(data = df_delta2,
paired = "baseline", id_col="ID",
x = ["Treatment", "Rep"], y = "Y",
delta2 = True, experiment = "Genotype")
paired_delta2.mean_diff.plot();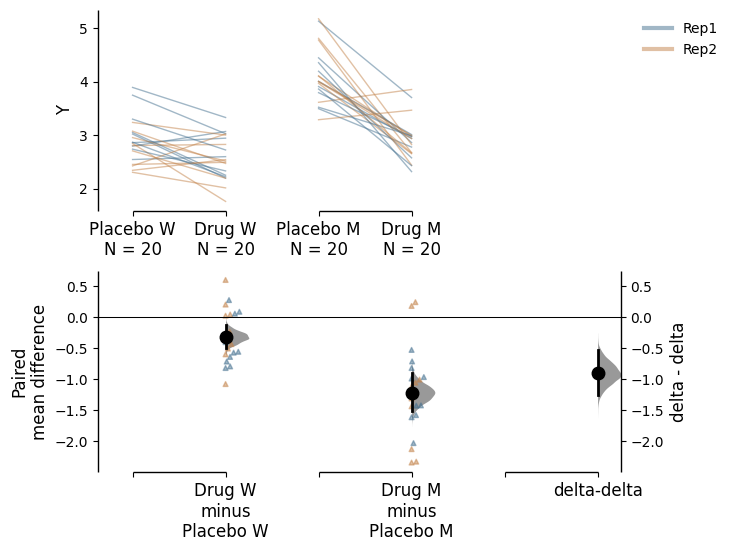
We see that the drug had a non-specific effect of -0.321 [95%CI -0.498, -0.131] on wild type subjects even when they were not sick, and it had a bigger effect of -1.22 [95%CI -1.52, -0.906] in mutant subjects. In this visualisation, we can see the delta-delta value of -0.903 [95%CI -1.21, -0.587] as the net effect of the drug accounting for non-specific actions in healthy individuals.
The mean difference between drug and placebo treatments in wild type subjects is:
\[\Delta_{1} = \overline{X}_{D, W} - \overline{X}_{P, W}\]
The mean difference between drug and placebo treatments in mutant subjects is:
\[\Delta_{2} = \overline{X}_{D, M} - \overline{X}_{P, M}\]
The net effect of the drug on mutants is:
\[\Delta_{\Delta} = \Delta_{2} - \Delta_{1}\]
where \(\overline{X}\) is the sample mean, \(\Delta\) is the mean difference.
Standardized mean difference statistics like Cohen’s d and Hedges’ g quantify effect sizes in terms of the sample variance. We have introduced a metric, Deltas’ g, to standardize delta-delta effects. This metric enables the comparison between measurements of different dimensions.
The standard deviation of the delta-delta value is calculated from a pooled variance of the 4 samples:
\[s_{\Delta_{\Delta}} = \sqrt{\frac{(n_{D, W}-1)s_{D, W}^2+(n_{P, W}-1)s_{P, W}^2+(n_{D, M}-1)s_{D, M}^2+(n_{P, M}-1)s_{P, M}^2}{(n_{D, W} - 1) + (n_{P, W} - 1) + (n_{D, M} - 1) + (n_{P, M} - 1)}}\]
where \(s\) is the standard deviation and \(n\) is the sample size.
A deltas’ g value is then calculated as delta-delta value divided by pooled standard deviation \(s_{\Delta_{\Delta}}\):
\(\Delta_{g} = \frac{\Delta_{\Delta}}{s_{\Delta_{\Delta}}}\)
DABEST v2024.03.29
==================
Good afternoon!
The current time is Tue Mar 19 15:44:15 2024.
The unpaired deltas' g between W Placebo and M Placebo is 2.54 [95%CI 1.68, 3.28].
The p-value of the two-sided permutation t-test is 0.0, calculated for legacy purposes only.
The unpaired deltas' g between W Drug and M Drug is 0.793 [95%CI 0.152, 1.34].
The p-value of the two-sided permutation t-test is 0.0122, calculated for legacy purposes only.
The deltas' g between Placebo and Drug is -2.11 [95%CI -2.97, -1.22].
The p-value of the two-sided permutation t-test is 0.0, calculated for legacy purposes only.
5000 bootstrap samples were taken; the confidence interval is bias-corrected and accelerated.
Any p-value reported is the probability of observing the effect size (or greater),
assuming the null hypothesis of zero difference is true.
For each p-value, 5000 reshuffles of the control and test labels were performed.
To get the results of all valid statistical tests, use `.delta_g.statistical_tests`We see the standardised delta-delta value of -2.11 standard deviations [95%CI -2.98, -1.2] as the net effect of the drug accounting for non-specific actions in healthy individuals.
The configuration of comparison we performed above is reminiscent of a two-way ANOVA. In fact, the delta - delta is an effect size estimated for the interaction term between Treatment and Genotype. Main effects of Treatment and Genotype, on the other hand, can be determined by simpler, univariate contrast plots.
If for some reason you don’t want to display the delta-delta plot, you can easily do so by
Since the delta-delta function is only applicable to mean differences, plots of other effect sizes will not include a delta-delta bootstrap plot.
You can find all outputs of the delta-delta calculation by assessing the attribute named delta_delta of the effect size object.
DABEST v2024.03.29
==================
Good afternoon!
The current time is Tue Mar 19 15:44:20 2024.
The delta-delta between Placebo and Drug is -0.903 [95%CI -1.27, -0.522].
The p-value of the two-sided permutation t-test is 0.0, calculated for legacy purposes only.
5000 bootstrap samples were taken; the confidence interval is bias-corrected and accelerated.
Any p-value reported is the probability of observing the effect size (or greater),
assuming the null hypothesis of zero difference is true.
For each p-value, 5000 reshuffles of the control and test labels were performed.DABEST v2024.03.29
==================
Good afternoon!
The current time is Tue Mar 19 15:44:20 2024.
The deltas' g between Placebo and Drug is -2.11 [95%CI -2.97, -1.22].
The p-value of the two-sided permutation t-test is 0.0, calculated for legacy purposes only.
5000 bootstrap samples were taken; the confidence interval is bias-corrected and accelerated.
Any p-value reported is the probability of observing the effect size (or greater),
assuming the null hypothesis of zero difference is true.
For each p-value, 5000 reshuffles of the control and test labels were performed.The delta_delta object has its own attributes, containing various information of delta - delta.
difference: the mean bootstrapped differences between the 2 groups of bootstrapped mean differencesbootstraps: the 2 groups of bootstrapped mean differencesbootstraps_delta_delta: the bootstrapped differences between the 2 groups of bootstrapped mean differencespermutations: the mean difference between the two groups of bootstrapped mean differences calculated based on the permutation datapermutations_var: the pooled group variances of two groups of bootstrapped mean differences calculated based on permutation datapermutations_delta_delta: the delta-delta calculated based on the permutation datadelta_delta.to_dict() will return all the attributes in a dictionary format.