import numpy as np
import pandas as pd
import dabest
print("We're using DABEST v{}".format(dabest.__version__))We're using DABEST v2024.03.29Since v2024.03.29, swarmplots are, by default, plotted asymmetrically to the right side. For detailed information, please refer to Swarm Side
import numpy as np
import pandas as pd
import dabest
print("We're using DABEST v{}".format(dabest.__version__))We're using DABEST v2024.03.29from scipy.stats import norm # Used in generation of populations.
np.random.seed(9999) # Fix the seed to ensure reproducibility of results.
Ns = 20 # The number of samples taken from each population
# Create samples
c1 = norm.rvs(loc=3, scale=0.4, size=Ns)
c2 = norm.rvs(loc=3.5, scale=0.75, size=Ns)
c3 = norm.rvs(loc=3.25, scale=0.4, size=Ns)
t1 = norm.rvs(loc=3.5, scale=0.5, size=Ns)
t2 = norm.rvs(loc=2.5, scale=0.6, size=Ns)
t3 = norm.rvs(loc=3, scale=0.75, size=Ns)
t4 = norm.rvs(loc=3.5, scale=0.75, size=Ns)
t5 = norm.rvs(loc=3.25, scale=0.4, size=Ns)
t6 = norm.rvs(loc=3.25, scale=0.4, size=Ns)
# Add a `gender` column for coloring the data.
females = np.repeat('Female', Ns/2).tolist()
males = np.repeat('Male', Ns/2).tolist()
gender = females + males
# Add an `id` column for paired data plotting.
id_col = pd.Series(range(1, Ns+1))
# Combine samples and gender into a DataFrame.
df = pd.DataFrame({'Control 1' : c1, 'Test 1' : t1,
'Control 2' : c2, 'Test 2' : t2,
'Control 3' : c3, 'Test 3' : t3,
'Test 4' : t4, 'Test 5' : t5, 'Test 6' : t6,
'Gender' : gender, 'ID' : id_col
})Use the parameter color_col to specify which column in the dataframe will be used to create the different colours for your graph.
The colour palette for the graph can be changed using the parameter custom_palette. All values from matplotlib or seaborn color palettes are accepted.
Additionally, a customized color palette can be defined by creating a dictionary where the keys are group names, and the values are valid matplotlib colours.
There are many ways to specify matplotlib colours. Find one example below using accepted colour names, hex strings (commonly used on the web), and RGB tuples.
By default, dabest.plot() desaturates the colour of the dots in the swarmplot by 50%. This draws attention to the effect size bootstrap curves.
You can alter the default values with the parameters swarm_desat and halfviolin_desat.
It is possible change the size of the dots used in the rawdata swarmplot, as well as those to indicate the effect sizes, by using the parameters raw_marker_size and es_marker_size respectively.
To change the y-limits for the rawdata axes, and the contrast axes, use the parameters swarm_ylim and contrast_ylim.
If the effect size is qualitatively inverted (ie. a smaller value is a better outcome), you can simply invert the tuple passed to contrast_ylim.
The contrast axes share the same y-limits as those of the delta-delta plot. Thus, the y axis of the delta-delta plot changes as well.
np.random.seed(9999) # Fix the seed so the results are replicable.
# Create samples
N = 20
y = norm.rvs(loc=3, scale=0.4, size=N*4)
y[N:2*N] = y[N:2*N]+1
y[2*N:3*N] = y[2*N:3*N]-0.5
# Add a `Treatment` column
t1 = np.repeat('Placebo', N*2).tolist()
t2 = np.repeat('Drug', N*2).tolist()
treatment = t1 + t2
# Add a `Rep` column as the first variable for the 2 replicates of experiments done
rep = []
for i in range(N*2):
rep.append('Rep1')
rep.append('Rep2')
# Add a `Genotype` column as the second variable
wt = np.repeat('W', N).tolist()
mt = np.repeat('M', N).tolist()
wt2 = np.repeat('W', N).tolist()
mt2 = np.repeat('M', N).tolist()
genotype = wt + mt + wt2 + mt2
# Add an `id` column for paired data plotting.
id = list(range(0, N*2))
id_col = id + id
# Combine all columns into a DataFrame.
df_delta2 = pd.DataFrame({'ID' : id_col,
'Rep' : rep,
'Genotype' : genotype,
'Treatment': treatment,
'Y' : y
})
paired_delta2 = dabest.load(data = df_delta2,
paired = "baseline", id_col="ID",
x = ["Treatment", "Rep"], y = "Y",
delta2 = True, experiment = "Genotype")
paired_delta2.mean_diff.plot(contrast_ylim=(3, -3),
contrast_label="More negative is better!");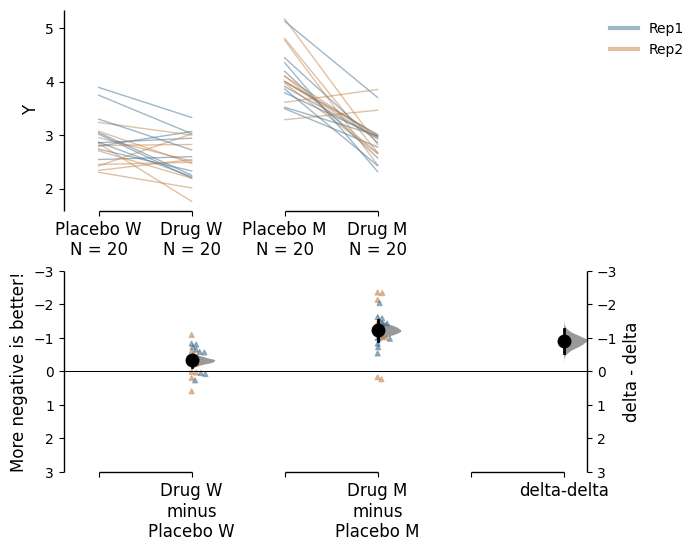
You can also change the `y-limits and y-label for the delta-delta plot.
You can add minor ticks and also change the tick frequency by accessing the axes directly.
Each estimation plot produced by dabest has two axes. The first one contains the rawdata swarmplot while the second one contains the bootstrap effect size differences.
import matplotlib.ticker as Ticker
f = two_groups_unpaired.mean_diff.plot()
rawswarm_axes = f.axes[0]
contrast_axes = f.axes[1]
rawswarm_axes.yaxis.set_major_locator(Ticker.MultipleLocator(1))
rawswarm_axes.yaxis.set_minor_locator(Ticker.MultipleLocator(0.5))
contrast_axes.yaxis.set_major_locator(Ticker.MultipleLocator(0.5))
contrast_axes.yaxis.set_minor_locator(Ticker.MultipleLocator(0.25))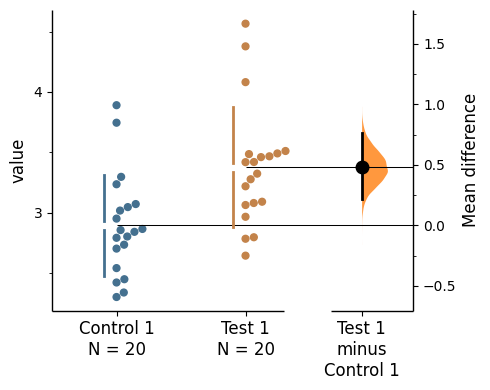
f = multi_2group.mean_diff.plot(swarm_ylim=(0,6),
contrast_ylim=(-3, 1))
rawswarm_axes = f.axes[0]
contrast_axes = f.axes[1]
rawswarm_axes.yaxis.set_major_locator(Ticker.MultipleLocator(2))
rawswarm_axes.yaxis.set_minor_locator(Ticker.MultipleLocator(1))
contrast_axes.yaxis.set_major_locator(Ticker.MultipleLocator(0.5))
contrast_axes.yaxis.set_minor_locator(Ticker.MultipleLocator(0.25))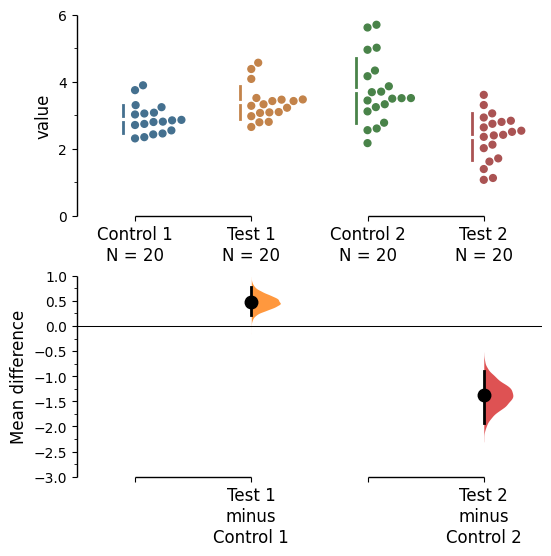
In dabest, swarmplots are, by default, plotted asymmetrically to the right side. You may change this by using the parameter swarm_side.
There are only three valid values: “right” (default), “left”, “center”.
For mini-meta plots, it is possible to hide the weighted average plot by setting the parameter show_mini_meta=False in the plot() function.
np.random.seed(9999) # Fix the seed so the results are replicable.
# pop_size = 10000 # Size of each population.
Ns = 20 # The number of samples taken from each population
# Create samples
c1 = norm.rvs(loc=3, scale=0.4, size=Ns)
c2 = norm.rvs(loc=3.5, scale=0.75, size=Ns)
c3 = norm.rvs(loc=3.25, scale=0.4, size=Ns)
t1 = norm.rvs(loc=3.5, scale=0.5, size=Ns)
t2 = norm.rvs(loc=2.5, scale=0.6, size=Ns)
t3 = norm.rvs(loc=3, scale=0.75, size=Ns)
# Add a `gender` column for coloring the data.
females = np.repeat('Female', Ns/2).tolist()
males = np.repeat('Male', Ns/2).tolist()
gender = females + males
# Add an `id` column for paired data plotting.
id_col = pd.Series(range(1, Ns+1))
# Combine samples and gender into a DataFrame.
df = pd.DataFrame({'Control 1' : c1, 'Test 1' : t1,
'Control 2' : c2, 'Test 2' : t2,
'Control 3' : c3, 'Test 3' : t3,
'Gender' : gender, 'ID' : id_col
})
mini_meta_paired = dabest.load(df, idx=(("Control 1", "Test 1"), ("Control 2", "Test 2"), ("Control 3", "Test 3")), mini_meta=True, id_col="ID", paired="baseline")
mini_meta_paired.mean_diff.plot(show_mini_meta=False);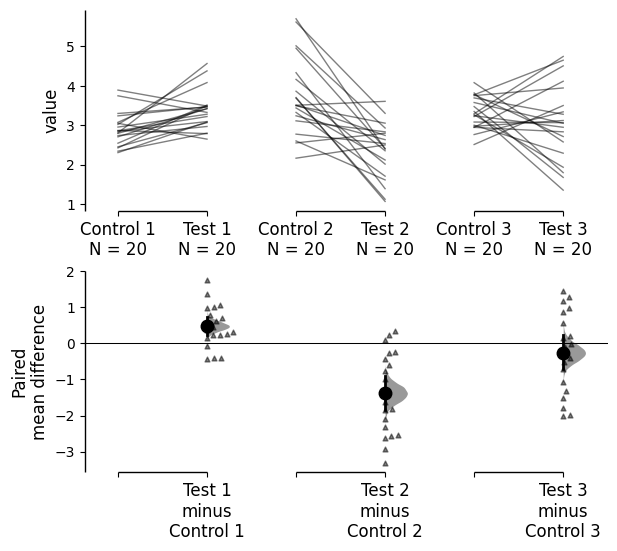
Similarly, you can also hide the delta-delta plot by setting show_delta2=False in the plot() function.
Implemented in v0.2.6 by Adam Nekimken.
dabest.plot has an ax parameter that accepts Matplotlib Axes. The entire estimation plot will be created in the specified Axes.
from matplotlib import pyplot as plt
f, axx = plt.subplots(nrows=2, ncols=2,
figsize=(15, 15),
gridspec_kw={'wspace': 0.25} # ensure proper width-wise spacing.
)
two_groups_unpaired.mean_diff.plot(ax=axx.flat[0]);
two_groups_paired_baseline.mean_diff.plot(ax=axx.flat[1]);
multi_2group.mean_diff.plot(ax=axx.flat[2]);
multi_2group_paired.mean_diff.plot(ax=axx.flat[3]);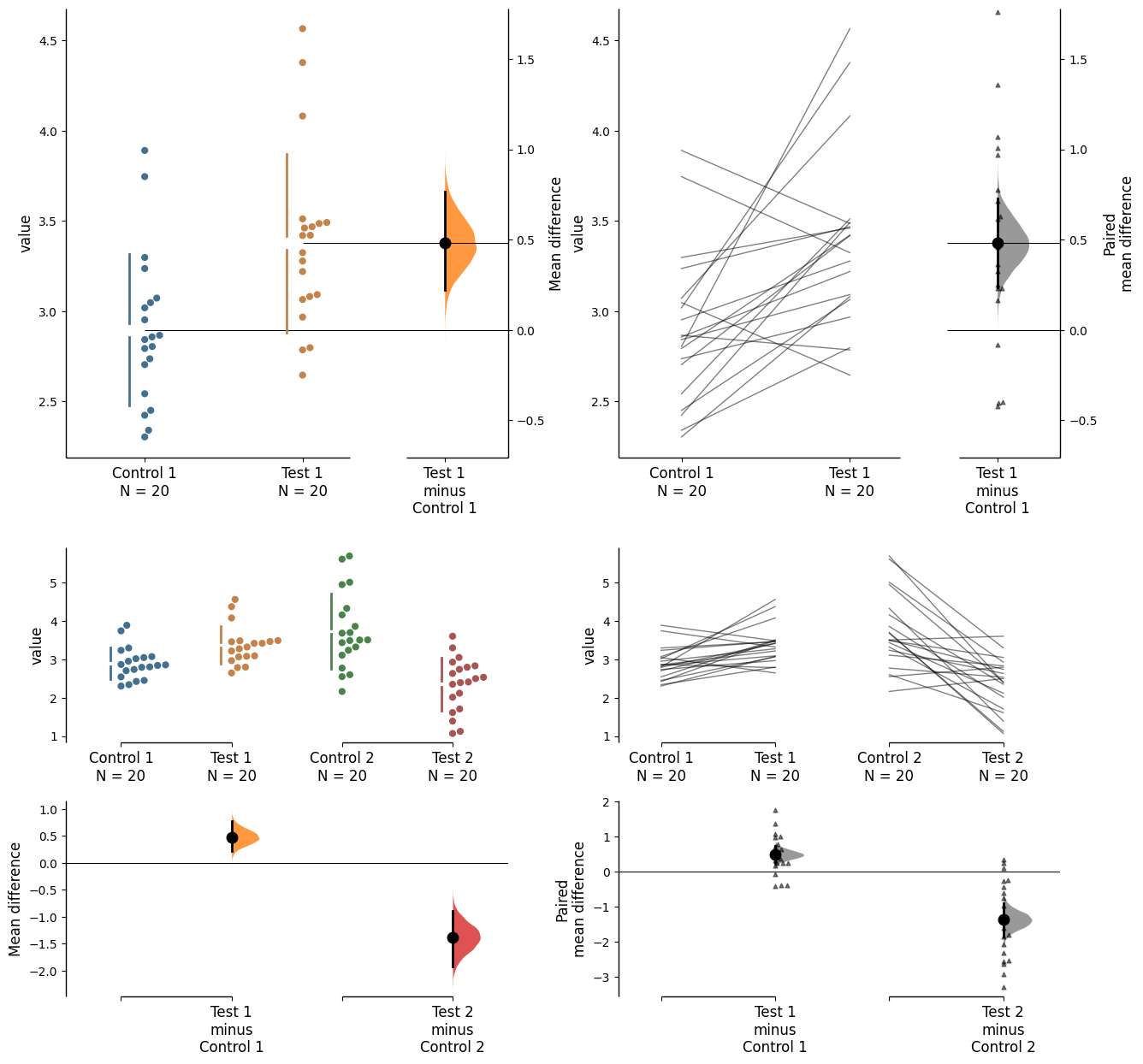
In this case, to access the individual rawdata axes, use name_of_axes to manipulate the rawdata swarmplot axes, and name_of_axes.contrast_axes to gain access to the effect size axes.